| Product name: |
Ref-1 (Acetyl Lys7) rabbit pAb |
| Reactivity: |
Human;Rat;Mouse; |
| Alternative Names: |
APEX1; APE; APE1; APEX; APX; HAP1; REF1; DNA-(apurinic or apyrimidinic site) lyase; APEX nuclease; APEN; Apurinic-apyrimidinic endonuclease 1; AP endonuclease 1; APE-1; REF-1; Redox factor-1 |
| Source: |
Rabbit |
| Dilutions: |
WB 1:500-2000;IHC-p 1:50-300 |
| Immunogen: |
The antiserum was produced against synthesized Acetyl-peptide derived from human APE1 around the Acetylation site of Lys7. AA range:1-50 |
| Storage: |
-20°C/1 year |
| Clonality: |
Polyclonal |
| Isotype: |
IgG |
| Concentration: |
1 mg/ml |
| Observed Band: |
35kD |
| GeneID: |
328 |
| Human Swiss-Prot No: |
P27695 |
| Cellular localization: |
Nucleus. Nucleus, nucleolus. Nucleus speckle. Endoplasmic reticulum. Cytoplasm. Detected in the cytoplasm of B-cells stimulated to switch (By similarity). Colocalized with SIRT1 in the nucleus. Colocalized with YBX1 in nuclear speckles after genotoxic stress. Together with OGG1 is recruited to nuclear speckles in UVA-irradiated cells. Colocalized with nucleolin and NPM1 in the nucleolus. Its nucleolar localization is cell cycle dependent and requires active rRNA transcription. Colocalized with calreticulin in the endoplasmic reticulum. Translocation from the nucleus to the cytoplasm is stimulated in presence of nitric oxide (NO) and function in a CRM1-dependent manner, possibly as a consequence of demasking a nuclear export signal (amino acid position 64-80). S-nitrosylation at Cys-93 and |
| Background: |
Apurinic/apyrimidinic (AP) sites occur frequently in DNA molecules by spontaneous hydrolysis, by DNA damaging agents or by DNA glycosylases that remove specific abnormal bases. AP sites are pre-mutagenic lesions that can prevent normal DNA replication so the cell contains systems to identify and repair such sites. Class II AP endonucleases cleave the phosphodiester backbone 5' to the AP site. This gene encodes the major AP endonuclease in human cells. Splice variants have been found for this gene; all encode the same protein. [provided by RefSeq, Jul 2008], |




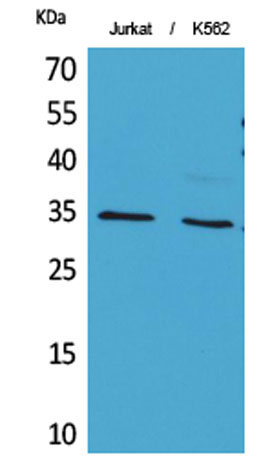 Western Blot analysis of Jurkat, K562 cells using Acetyl-Ref-1 (K7) Polyclonal Antibody. Secondary antibody(catalog#:RS0002) was diluted at 1:20000
Western Blot analysis of Jurkat, K562 cells using Acetyl-Ref-1 (K7) Polyclonal Antibody. Secondary antibody(catalog#:RS0002) was diluted at 1:20000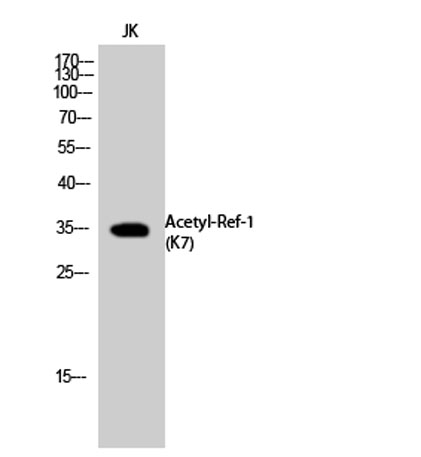 Western Blot analysis of JK cells using Acetyl-Ref-1 (K7) Polyclonal Antibody. Secondary antibody(catalog#:RS0002) was diluted at 1:20000
Western Blot analysis of JK cells using Acetyl-Ref-1 (K7) Polyclonal Antibody. Secondary antibody(catalog#:RS0002) was diluted at 1:20000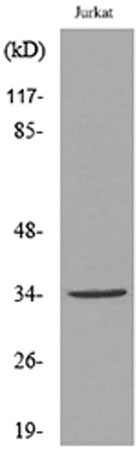 Western blot analysis of lysate from Jurkat cells, using APE1 (Acetyl-Lys7) Antibody.
Western blot analysis of lysate from Jurkat cells, using APE1 (Acetyl-Lys7) Antibody.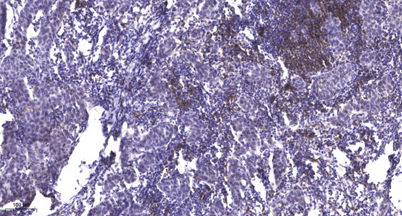 Immunohistochemical analysis of paraffin-embedded human Breast cancer. 1, Antibody was diluted at 1:200(4° overnight). 2, Tris-EDTA,pH9.0 was used for antigen retrieval. 3,Secondary antibody was diluted at 1:200(room temperature, 45min).
Immunohistochemical analysis of paraffin-embedded human Breast cancer. 1, Antibody was diluted at 1:200(4° overnight). 2, Tris-EDTA,pH9.0 was used for antigen retrieval. 3,Secondary antibody was diluted at 1:200(room temperature, 45min).在線咨詢
技術(shù)支持


 下載說明 ①
下載說明 ①



